REgulating Active DNA MEthylation (README)
Clara Cena
Aree / Gruppi di ricerca
Partecipanti al progetto
- Bertinaria Massimo (Docente)
- Cena Clara (Docente)
- Costale Annalisa (Tecnico/a)
- Giorgis Marta (Ricercatore/trice)
- Marini Elisabetta (Ricercatore/trice)
Descrizione del progetto

Sponsors:
University of Turin
Description
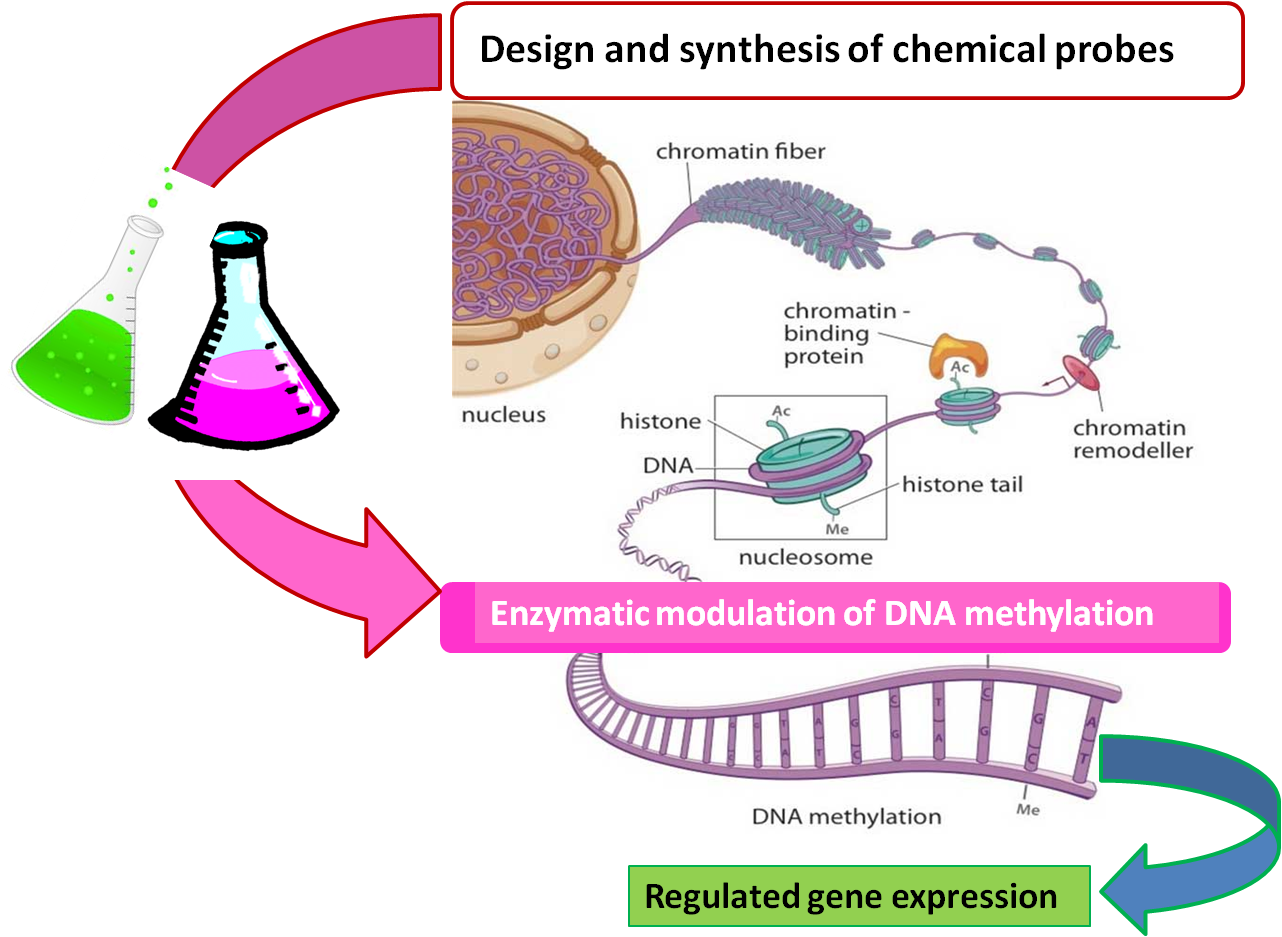
Design and synthesis on new chemical entities modulating the activity of DNA methyl transferases (DNMTs) and Ten-Eleven Translocation factor proteins.
DNA methylation is an epigenetic mark which plays an essential role in mammalian cell regulation.
DNA methylation patterns are established and maintained by several methyltransferase enzymes (DNMTs) and recent studies have shown that ten-eleven translocation proteins (TETs) can be involved in active DNA demethylation.
Alterations in DNA methylation patterns are associated with many different diseases, such as cancer, diabetes and cardiovascular pathologies, so both DNMT and TET enzymes are attractive targets for drug discovery.
The goal of this project is to identify new epigenetic modulators through design, synthesis and biological screening of small molecules.
Selected leads will be consequently modulated to obtain potent and selective new epigenetic drugs, able to interfere with the DNA methylation for therapeutic applications as gene expression regulators.
The unit is part of European COST action TD0905 “epigenetics bench to bedside” WG1.
Keywords:
DNMTs, TET, Gene expression, Enzyme modulators, Epigenetics, DNA




